Cas9 fusion protein and its coding sequence
A technology of fusion protein and coding sequence, which is applied in the field of DNA sequence, can solve the problems affecting gene targeting efficiency and chimeric effect, and achieve the effects of reducing gene targeting chimeric effect, improving the accuracy of gene modification, and reducing the effect of chimeric mutation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
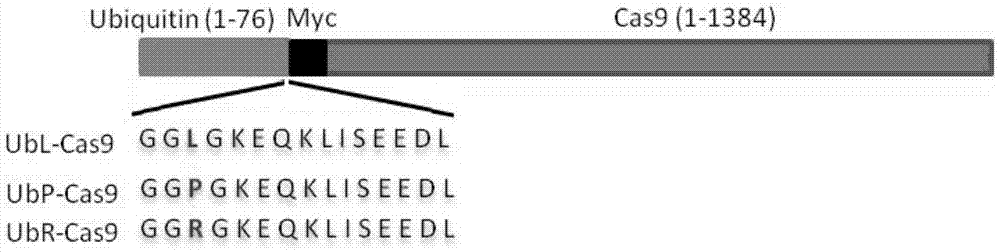
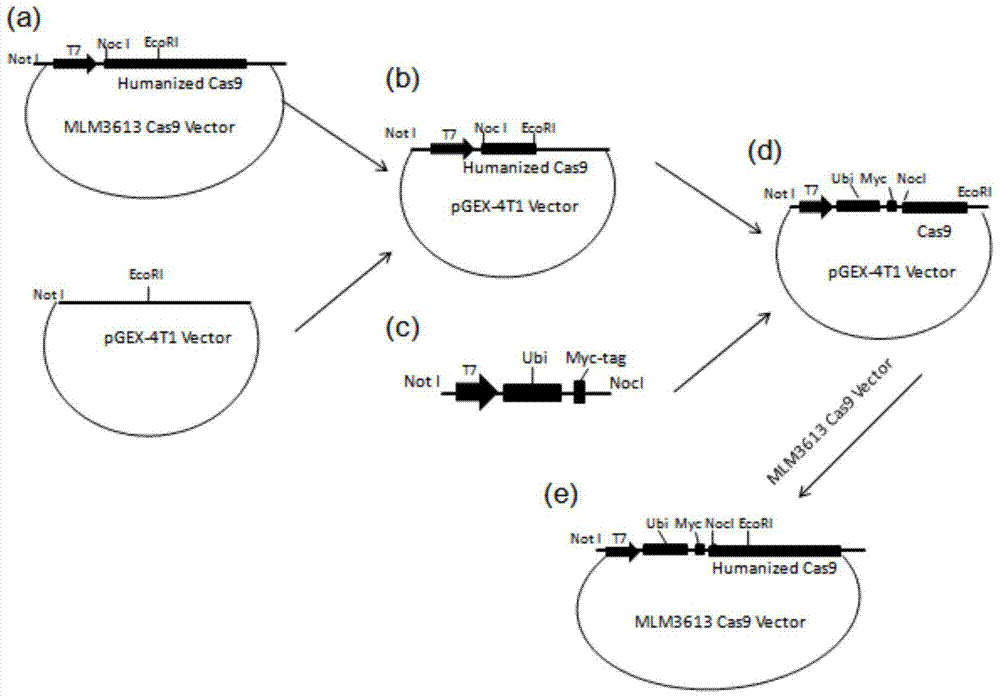
[0041] Construct the expression vector of the target plasmid Ub-i-Cas9 (i represents R, L, P) containing different protein degradation signals, the schematic diagram is as follows figure 1 , the construction process method is as follows ( figure 2 ).
[0042] For ease of construction, first use the NotI and EcoRI restriction sites on the MLM3613Cas9 (Addgene, Plasmid#42251, called WT-Cas9) backbone plasmid to contain the T7 promoter, NcoI cloning restriction site, and translation initiation site. Points and fragments of part of the Cas9 protein sequence were cut and ligated on the pGEX4T-1 plasmid ( figure 2 a, b), pGEX4T1-NotI-T7-NcoI-Cas9-EcoRI-. Subsequently, design upstream primers with NotI restriction site and T7 promoter sequence I and downstream primers with Myc-tag sequence, L / P / L site and NocI restriction site (sequences are respectively as SEQ ID NO .22, shown in SEQ ID NO.23-25), introduce the DNA sequence of L / P / R amino acid simultaneously in the downstream p...
Embodiment 2
[0056] Construction of CRISPR / Cas9 sgRNA vectors containing PINK1, Parkin and ASPM gene target sequences.
[0057] Table 1. Target Sequence Sites
[0058]
[0059] Note: The sequence underlined is the PAM sequence of the sgRNA, that is, its binding site with the target DNA sequence.
[0060] The sgRNA target sequence vector constructed above and the Cas9 plasmid vectors before and after transformation were linearized with PmeI enzyme, recovered from the gel, and synthesized by in vitro transcription.
[0061]The sgRNA expression vector contains a T7 promoter to control the expression of sgRNA, and can insert a 20bp sgRNA specific target sequence after digestion with BbsI restriction enzyme, and the recognition site of the specific target sequence is located in the second exon of PINK1 gene , the second exon of the Parkin gene, the third exon and the tenth exon of the ASPM gene. In order to screen the sgRNA sequences of PINK1 and ASPM genes capable of gene editing and with...
Embodiment 3
[0063] The half-life detection of the constructed Ub-i-Cas9 (i stands for LP, R) plasmid.
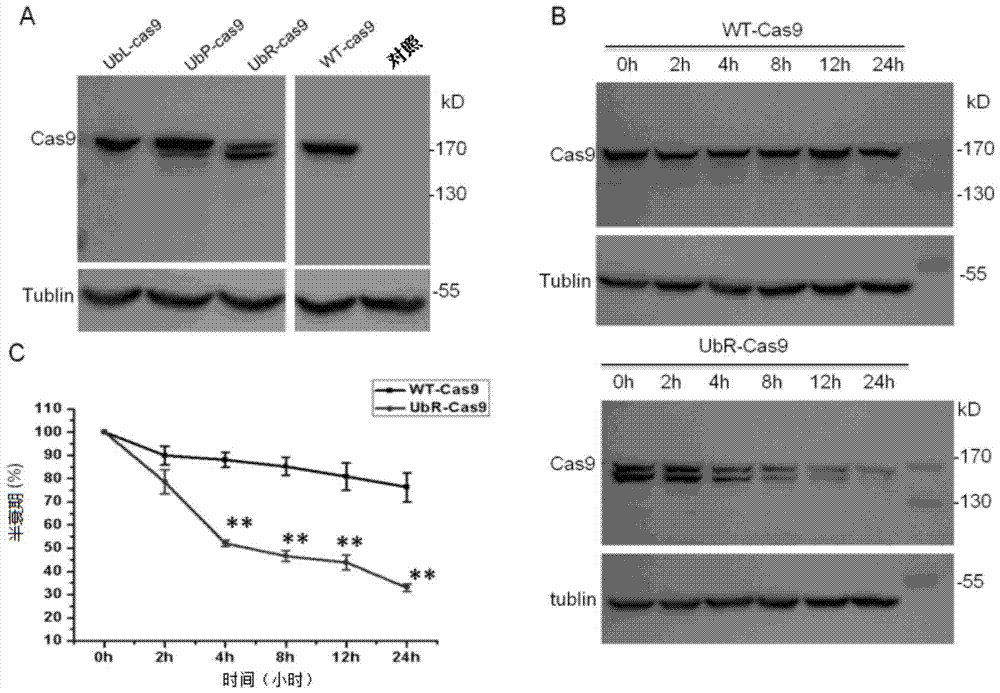
[0064] First, WT-Cas9 and Ub-i-Cas9 plasmids were transfected into 293 cells. After 48 hours of transfection, the results of Western blot detection of Cas9 protein expression showed that, based on WT-Cas9 as 100%, Ub-L-Cas9, Ub-L-Cas9, Ub-P-Cas9 and Ub-R-Cas9 were degraded to 84.76%, 80.92% and 63.89%, respectively.
[0065] Especially for Ub-R-Cas9, although there is only one base difference between Ub-L-Cas9 and Ub-P-Cas9 in its sequence, it is surprisingly found that its degradation rate is significantly faster than other fusion proteins, reached a very rare level.
[0066] Subsequently, we compared the half-life of WT-Cas9 and Ub-R-Cas9, that is, after 24 hours of plasmid transfection, 50 micromolar cycloheximide was added to the medium, respectively at 0h, 2h, 4h, 8h, 12h , 24h to collect cells, Western blot detection results are as follows image 3 As shown in B and C, compared...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap